Single-nucleotide polymorphisms (SNPs) are the most common type of genetic variation among people, with an average of one SNP per 1,000 nucleotides. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. They can also be used to track the inheritance of disease-associated genetic variants within families. SNPs are used in genome-wide association studies (GWAS) to identify regions of the genome that may cause common complex diseases. These studies have led to the development of genetic counselling and improved individual disease risk assessment. SNPs are also used in disease gene mapping, medicinal drug development, and understanding human genome evolution.
| Characteristics | Values |
|---|---|
| Definition | Single-nucleotide polymorphism (SNP) is a germline substitution of a single nucleotide at a specific position in the genome. |
| Occurrence | SNPs occur almost once in every 1,000 nucleotides on average, which means there are roughly 4 to 5 million SNPs in a person's genome. |
| Function | SNPs can be used to identify disease-causing genes, predict an individual's response to certain drugs, susceptibility to environmental factors, and risk of developing diseases. |
| Use Cases | SNPs have been used in forensics, pharmacogenetics, disease causation, and biomedical research. |
| Databases | dbSNP, Kaviar, SNPedia, OMIM, dbSAP, The Human Gene Mutation Database, GWAS Central |
| Limitations | SNPs yield less information than STRs, and therefore more SNPs are needed for analysis. They also rely on the presence of a database for comparative analysis. |
Explore related products
$126.7 $179
What You'll Learn
- SNPs are the most common type of genetic variation among people
- SNPs can be used to identify disease-causing genes
- SNPs can help explain differences in susceptibility to diseases
- SNPs can be used to track the inheritance of disease-associated genetic variants
- SNPs can be used to understand the molecular mechanisms of sequence evolution

SNPs are the most common type of genetic variation among people
Single-nucleotide polymorphisms, or SNPs, are the most common type of genetic variation among people. Each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA. SNPs occur normally throughout a person's DNA, with an average frequency of about one SNP per 1,000 nucleotides. This means there are roughly 4 to 5 million SNPs in a person's genome.
SNPs are classified as such when a variant is found in at least 1% of the population. However, this frequency threshold is not always applied in publications. SNPs are most commonly found in the DNA between genes. They can act as biological markers, helping scientists locate genes that are associated with diseases. SNPs are also used to track the inheritance of disease-associated genetic variants within families.
SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the CFH gene is associated with an increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For instance, two common SNPs in the APOE gene, rs429358 and rs7412, lead to three major APO-E alleles with different associated risks for the development of Alzheimer's disease and age at onset.
SNPs are also used in biomedical research, forensics, pharmacogenetics, and disease causation. Historically, SNPs were used to match a forensic DNA sample to a suspect, but this technique has become obsolete due to advancing STR-based DNA fingerprinting techniques. However, the development of next-generation sequencing (NGS) technology may allow for more opportunities to use SNPs in phenotypic clues such as ethnicity, hair colour, and eye colour, with a good probability of a match.
Spanish Skills: Border Patrol's Reasonable Suspicion
You may want to see also

SNPs can be used to identify disease-causing genes
Single-nucleotide polymorphisms (SNPs) are the most common type of genetic variation among people. Each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with thymine (T) in a certain stretch of DNA. SNPs occur naturally and frequently throughout the human genome, about once in every 1,000 nucleotides on average, translating to roughly 4 to 5 million SNPs in a person's genome.
SNPs can also be used to track the inheritance of disease-associated genetic variants within families and to study sequence variation among species. They are also critical for personalised medicine, including biomedical research, forensics, pharmacogenetics, and disease causation. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the CFH gene is associated with an increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs.
Additionally, SNPs can be used to understand the molecular mechanisms of sequence evolution. Natural selection will maintain the amino acid type and retain the amino acid position among species because these amino acids are critical to protein function. Deleterious mutations that affect the biological functions of proteins are effectively eliminated by natural selection from the gene pool. As a result, a comparative genomic study of disease-associated SNPs can be used to understand the relationship between pathology and evolution.
Child Abuse: Constitutional Rights and Wrongs
You may want to see also

SNPs can help explain differences in susceptibility to diseases
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation among people. They occur almost once in every 1,000 nucleotides on average, meaning there are roughly 4 to 5 million SNPs in a person's genome. SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the CFH gene is associated with an increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For instance, two common SNPs in the APOE gene, rs429358 and rs7412, lead to three major APO-E alleles with different associated risks for the development of Alzheimer's disease and the age of onset.
SNPs can also be used to predict an individual’s response to certain drugs, susceptibility to environmental factors such as toxins, and the risk of developing diseases. They can be used to track the inheritance of disease-associated genetic variants within families. Research is ongoing to identify SNPs associated with complex diseases such as heart disease, diabetes, and cancer. SNPs in non-coding regions can manifest in a higher risk of cancer, affect mRNA structure, and disease susceptibility. Non-coding SNPs can also alter the level of gene expression.
In genetic epidemiology, SNPs are used to estimate transmission clusters. Variations in the DNA sequences of humans can affect how humans develop diseases and respond to pathogens, chemicals, drugs, vaccines, and other agents. SNPs are also critical for personalized medicine and are used in biomedical research, forensics, pharmacogenetics, and disease causation. One of the main contributions of SNPs in clinical research is the genome-wide association study (GWAS). GWAS has been commonly used to identify SNPs associated with diseases or clinical phenotypes or traits.
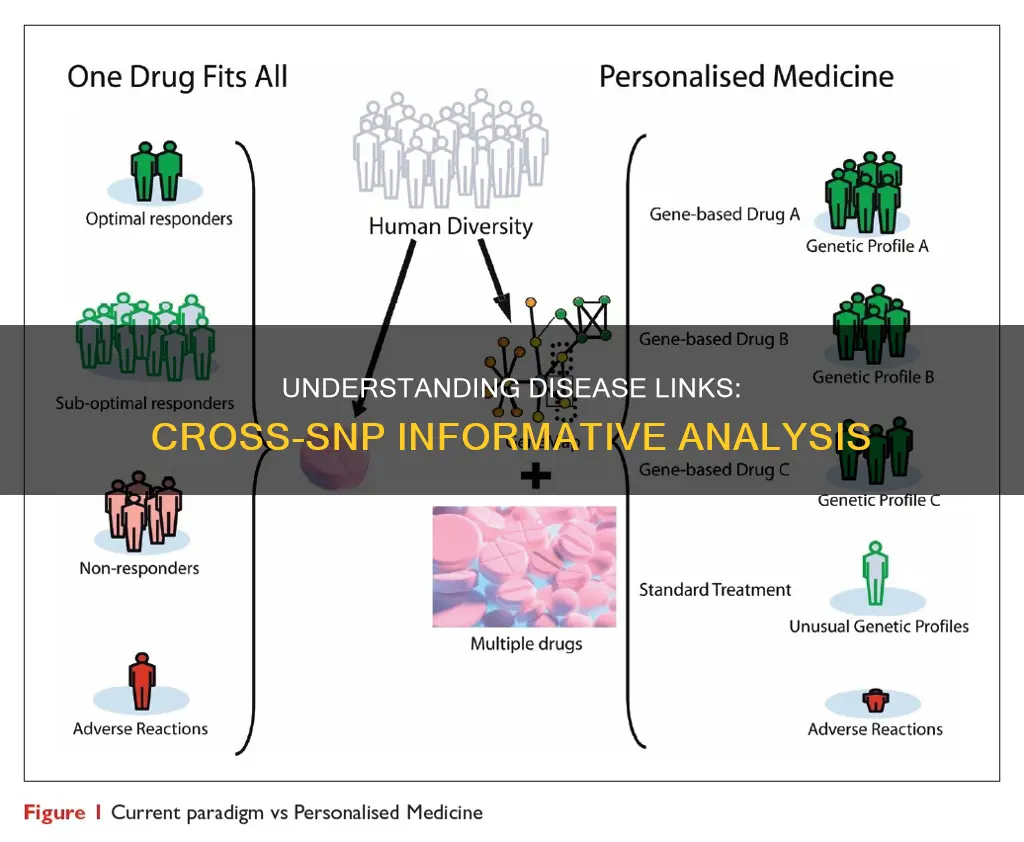
SNPs are also used in disease gene mapping and medicinal drug development. They can be used to identify disease-causing genes in humans and to understand the inter-individual variation in drug response. By establishing an association between the genetic makeup of an individual and their drug response, it may be possible to develop a genome-based diet and medicines that are more effective and safer for each individual.
Judicial Review: Upholding the Constitution's Principles and Values
You may want to see also
Explore related products

SNPs can be used to track the inheritance of disease-associated genetic variants
Single-nucleotide polymorphisms (SNPs) are the most common type of genetic variation among people. Each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA. SNPs occur almost once in every 1,000 nucleotides on average, which means there are roughly 4 to 5 million SNPs in a person's genome.
SNPs can help explain differences in susceptibility to a wide range of diseases across a population. For example, a common SNP in the CFH gene is associated with an increased risk of age-related macular degeneration. Differences in the severity of an illness or response to treatments may also be manifestations of genetic variations caused by SNPs. For instance, two common SNPs in the APOE gene, rs429358 and rs7412, lead to three major APO-E alleles with different associated risks for the development of Alzheimer's disease and the age of onset.
SNPs are also used in association studies to map disease-causing mutations. Because SNPs have only two alleles, they can be genotyped by a simple plus/minus assay rather than a length measurement, making them more amenable to automation. This type of analysis, called haplotype analysis, has been used to identify common disease genes.
The British Constitution: A Complex Web of Documents
You may want to see also

SNPs can be used to understand the molecular mechanisms of sequence evolution
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation among people. They occur almost once in every 1,000 nucleotides on average, translating to roughly 4 to 5 million SNPs in a person's genome. SNPs can be used to understand the molecular mechanisms of sequence evolution.
SNPs can be used to identify disease-causing genes in humans and to understand inter-individual variation in drug response. By establishing an association between an individual's genetic makeup and their response to drugs, it may be possible to develop more effective and safer treatments. SNPs can also be used to track the inheritance of disease-associated genetic variants within families. For example, two common SNPs in the APOE gene, rs429358 and rs7412, are associated with different risks for the development of Alzheimer's disease and the age of onset.
Additionally, SNPs can be used to understand the rate, type, and site of nucleotide substitutions, as well as the selection pressure on codons. Natural selection maintains and retains the amino acid type and position among species as these amino acids are critical for protein function. Deleterious mutations that affect the biological functions of proteins are effectively eliminated by natural selection.
SNPs can also be used to study the evolution of conserved residues, which are evolving under strong selective pressure. By comparing the relative fixation rates of silent and non-synonymous SNPs, researchers can trace the branch point of an evolutionary tree, where the variants became advantageous for the species and fixed in the gene pool.
Furthermore, new molecular genetic techniques involving SNPs have impacted the fields of ecology, evolution, and conservation. SNPs can be used as genetic markers to answer questions in population genetics, providing broader genome coverage and higher-quality data than other markers. They can also be used to analyze population structures, gene flow, and gene migration by observing allele frequencies within a population.
When to Avoid the ER: Non-Emergency Situations
You may want to see also
Frequently asked questions
A single-nucleotide polymorphism (SNP) is a germline substitution of a single nucleotide at a specific position in the genome. They are the most common type of genetic variation among people, occurring once in every 1,000 nucleotides on average. SNPs can help explain differences in susceptibility to a wide range of diseases across a population.
SNPs are used to identify disease-causing genes in humans and to understand inter-individual variation in drug response. They are used in association studies to map disease-causing mutations and identify regions of the genome that may cause complex diseases. SNPs can also be used to track the inheritance of disease-associated genetic variants within families.
SNPs have been used to identify common disease genes and understand the molecular mechanisms of diseases such as migraine. For example, common SNPs related to migraine have helped understand the role of the glutamatergic system and cortical spreading depression in the pathogenesis of migraine. SNPs have also been used to identify genes involved in determining complex traits and to develop genetic counselling for improved individual disease risk assessment.











































